Computational Imaging is an interdisciplinary Working Group between TUM Neuroradiology and UZH DQBM, jointly headed by Dr. Benedikt Wiestler and Prof. Bjoern Menze.
Medical Imaging generates a plethora of data, of which today only a fraction is used for clinical decision-making. Within our Working Group, we aim to develop algorithms and strategies to make the wealth of information accessible to clinicians. To this end, we are developing tools for (un)supervised lesion detection / segmentation, classification, and data integration. Together with our clinical partners @ TUM, our current focus is on two neurological model diseases: Multiple Sclerosis and Gliomas. To support the dissemination and use of our results, we aim to make all tools developed by us available here. We also actively contribute to important challenges (BraTS) and workshops (BrainLes) in medical image computing.
News
- Marie Metz wins the 2023 Marc-Dünzl-Preis @ DGNR. Congratulations!
- Our joint project “Graph Learning and Pathophysiological Models: Towards a New Classification of White Matter Lesions in Multiple Sclerosis” (with TUM-AIM and TUM Neurology) is part of the second funding period of the DFG Priority Programme Radiomics.
- Our joint project “Improving risk stratification and treatment planning in brain metastases patients: Development and validation of AI models in the AURORA multi-center patient cohort” (with TUM-AIM and TUM Radiation Oncology) is funded by the DFG.
- Florian Kofler wins the Valdo Grand Challenge @ MICCAI. Congrats!
- Marie Metz receives a TUM-KKF scholarship to explore the clinical utility of tumor growth maps.
- Our joint project “DIVA” (with TUM-CAMP, Smart Reporting and ImFusion), aiming to develop an explainable decision-support system for detecting and classifying vertebral fractures, is funded by the KMU-innovativ programme of the BMBF.
- Bjoern Menze is joining the Department of Quantitative Biomedicine of UZH to establish the new Chair of Biomedical Image Analysis and Machine Learning.
- Tom Finck receives a TUM-KKF scholarship.
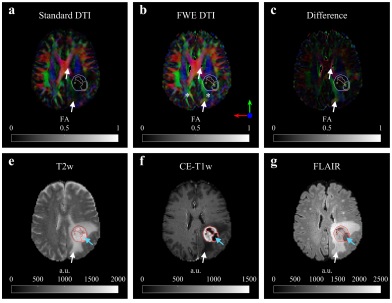
- Marie Metz wins a Trainee Research Award Neuroradiology @ RSNA 2019 for her work “Predicting Glioblastoma Recurrence from Preoperative MR Scans using Fractional Anisotropy Maps with Free Water Suppression”.
- Benedikt Wiestler is awarded the 2019 Kurt-Decker-Preis @ DGNR.
- Our proposal “Predicting individual disease activity in Multiple Sclerosis: Making the informative wealth of white matter lesion imaging clinically accessible” is selected to be funded in the DFG Priority Programme Radiomics.
People
Group Leaders
- Dr. Benedikt Wiestler (TUM Neuroradiology)
- Prof. Bjoern Menze (TUM IBBM & UZH DQBM)
Scientists
- PD Dr. Tom Finck (TUM Neuroradiology)
- Dr. Marie Metz (TUM Neuroradiology)
- Dr. Sarah Schlaeger (TUM Neuroradiology)
- Bailiang Jian (PhD Student)
- Hongwei Li (PhD Student)
- Daniel Scholz (PhD Student)
- Aswathi Varma (PhD Student)
- Jonas Weidner (PhD Student)
- Marie Thomas (MD Student)
Collaboration Partners
- Dr. Shadi Albarqouni (Helmholtz Munich / Uni Bonn)
- Prof. Spyridon Bakas (UPenn)
- Prof. Stephanie Combs (TUM Radiation Oncology)
- Dr. Claire Delbridge (TUM Neuropathology)
- Prof. Jens Gempt (TUM / Hamburg Neurosurgery)
- Prof. Jan Kirschke (TUM Neuroradiology)
- Prof. Mark Mühlau (TUM Neurology)
- Prof. Nassir Navab (TUM CAMP)
- Dr. Franz Pfister (deepc GmbH)
- Dr. Viola Pongratz (TUM Neurology)
- Prof. Alexander Radbruch (Bonn Neuroradiology)
- Prof. Daniel Rückert (TUM AIM)
- Prof. Peter Schüffler (TUM Computational Pathology)
- Prof. Philipp Vollmuth (Heidelberg Neuroradiology)
- Prof. Christian Wachinger (TUM Radiology)
- Prof. Roland Wiest (Bern Neuroradiology)
Selected Projects
 |
 |
 |
 |
 |
 |
Selected Publications
McGinnis J, Shit S, Li H, Sideri-Lampretsa V, Graf R, Dannecker M, Pan J, Stolt-Ansó N, Mühlau M, Kirschke J, Rueckert D, Wiestler B. Single-subject Multi-contrast MRI Super-resolution via Implicit Neural Representations. MICCAI 2023
Prabhakar C, Li H, Paetzold J, Loehr T, Niu C, Mühlau M, Rueckert D, Wiestler B*, Menze BH*. Self-pruning Graph Neural Network for Predicting Inflammatory Disease Activity in Multiple Sclerosis from Brain MR Images. MICCAI 2023
Prabhakar C, Li H, Yang J, Shit S, Wiestler B*, Menze BH*. ViT-AE++: Improving Vision Transformer Autoencoder for Self-supervised Medical Image Representations. MIDL, 2023
Kofler F, Shit S, Ezhov I, Fidon L, Horvath I, Al-Maskari R, Li H, Bhatia H, Loehr T, Piraud M, Ertuerk A, Kirschke J, Peeken J, Vercauteren T, Zimmer C, Wiestler B*, Menze BH*. blob loss: instance imbalance aware loss functions for semantic segmentation. IPMI, 2023
Ezhov I, Scibilia K, Franitza K, Steinbauer F, Shit S, Zimmer L, Lipkova J, Kofler F, Paetzold JC, Canalini L, Waldmannstetter D, Menten M, Metz M, Wiestler B*, Menze BH*. Learn-Morph-Infer: A new way of solving the inverse problem for brain tumor modeling. Medical Image Analysis, 2022
Bercea CI, Wiestler B, Rueckert D, Albarqouni S. Federated disentangled representation learning for unsupervised brain anomaly detection. Nature Machine Intelligence, 2022
Gempt J, Withake F, Aftahy AK, Meyer HS, Barz M, Delbridge C, Liesche-Starnecker F, Prokop G, Pfarr N, Schlegel J, Meyer B, Zimmer C, Menze BH*, Wiestler B*. Methylation subgroup and molecular heterogeneity is a hallmark of glioblastoma: implications for biopsy targeting, classification and therapy. ESMO Open, 2022
Ezhov I, Mot T, Shit S, Lipkova J, Paetzold JC, Kofler F, Pellegrini C, Kollovieh M, Navarro F, Li H, Metz M, Wiestler B, Menze BH. Geometry-aware neural solver for fast Bayesian calibration of brain tumor models. IEEE TMI, 2021
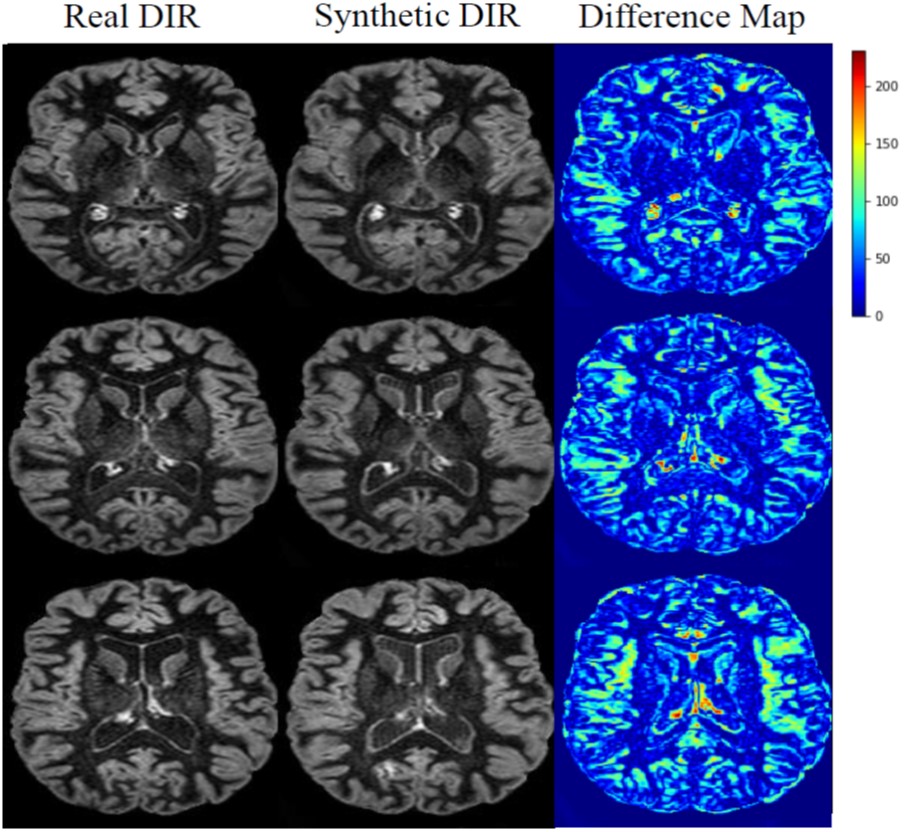
Thomas MF, Kofler F, Grundl L, Finck T, Li H, Zimmer C, Menze BH, Wiestler B. Improving Automated Glioma Segmentation in Routine Clinical Use Through Artificial Intelligence-Based Replacement of Missing Sequences With Synthetic Magnetic Resonance Imaging Scans. Investigative Radiology, 2021
Paprottka KJ, Kleiner S, Preibisch C, Kofler F, Schmidt-Graf F, Delbridge C, Bernhardt D, Combs SE, Gempt J, Meyer B, Zimmer C, Menze BH, Yakushev I, Kirschke JS, Wiestler B. Fully automated analysis combining 18F-FET-PET and multiparametric MRI including DSC perfusion and APTw imaging: a promising tool for objective evaluation of glioma progression. EJNMMI, 2021
Finck T, Schinz D, Grundl L, Eisawy R, Yigitsoy M, Moosbauer J, Pfister F, Wiestler B. Automated Pathology Detection and Patient Triage in Routinely Acquired Head Computed Tomography Scans. Investigative Radiology, 2021
Baur C*, Wiestler B*, Mühlau M, Zimmer C, Navab N, Albarqouni S. Modeling Healthy Anatomy with Artificial Intelligence for Unsupervised Anomaly Detection in Brain MRI. Radiology AI, 2021
Metz MC, Molina-Romero M, Lipkova J, Gempt J, Liesche-Starnecker F, Eichinger P, Grundl L, Menze B, Combs SE, Zimmer C, Wiestler B. Predicting Glioblastoma Recurrence from Preoperative MR Scans Using Fractional-Anisotropy Maps with Free-Water Suppression. Cancers, 2020
Li H, Paetzold J, Sekuboyina A, Kofler F, Zhang J, Kirschke JS, Wiestler B, Menze BH. DiamondGAN: Unified Multi-Modal Generative Adversarial Networks for MRI Sequences Synthesis. MICCAI, 2019
Baur C, Wiestler B, Albarqouni S, Navab N. Fusing Unsupervised and Supervised Deep Learning for White Matter Lesion Segmentation. MIDL, 2019
Eichinger P, Schön S, Pongratz V, Wiestler H, Zhang H, Bussas M, Hoshi MM, Kirschke JS, Berthele A, Zimmer C, Hemmer B, Mühlau M, Wiestler B. Accuracy of Unenhanced MRI in the Detection of New Brain Lesions in Multiple Sclerosis. Radiology, 2019
Lipkova J, Angelikopoulos P, Wu S, Alberts E, Wiestler B, Diehl C, Preibisch C, Pyka T, Combs S, Hadjidoukas P, Van Leemput K, Koumoutsakos P, Lowengrub JS, Menze BH. Personalized Radiotherapy Design for Glioblastoma: Integrating Mathematical Tumor Models, Multimodal Scans and Bayesian Inference. IEEE TMI, 2019
Molina-Romero M, Wiestler B, Gómez PA, Menzel MI, Menze BH. Deep Learning with Synthetic Diffusion MRI Data for Free-Water Elimination in Glioblastoma Cases. MICCAI, 2018
Zhang H, Alberts E, Pongratz V, Mühlau M, Zimmer C, Wiestler B, Eichinger P. Predicting conversion from clinically isolated syndrome to multiple sclerosis–An imaging-based machine learning approach. NeuroImage: Clinical, 2018
Baur C, Wiestler B, Albarqouni S, Navab N. Deep Autoencoding Models for Unsupervised Anomaly Segmentation in Brain MR Images. arXiv:1804.04488, 2018
Eichinger P, Wiestler H, Zhang H, Biberacher V, Kirschke JS, Zimmer C, Mühlau M, Wiestler B. A novel imaging technique for better detecting new lesions in multiple sclerosis. J Neurol, 2017
Eichinger P, Alberts E, Delbridge C, Trebeschi S, Valentinitsch A, Bette S, Huber T, Gempt J, Meyer B, Schlegel J, Zimmer C, Kirschke JS, Menze BH, Wiestler B. Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Scientific Reports, 2017
Alberts E, Tetteh G, Trebeschi S, Bieth M, Valentinitsch A, Wiestler B, Zimmer C, Menze BH. Multi-modal Image Classification Using Low-Dimensional Texture Features for Genomic Brain Tumor Recognition. MICGen @ MICCAI 2017
Menze BH, Van Leemput K, Lashkari D, Riklin-Raviv T, Geremia E, Alberts E, Gruber P, Wegener S, Weber MA, Székely G, Ayache N, Golland P. A Generative Probabilistic Model and Discriminative Extensions for Brain Lesion Segmentation - With Application to Tumor and Stroke. IEEE Trans. Med. Imaging, 2016
Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, Huang L, Ratliff M, Karimian Jazi K, Kurz FT, Schmenger T, Lemke D, Gömmel M, Pauli M, Liao Y, Häring P, Pusch S, Herl V, Steinhäuser C, Krunic D, Jarahian M, Miletic H, Berghoff AS, Griesbeck O, Kalamakis G, Garaschuk O, Preusser M, Weiss S, Liu H, Heiland S, Platten M, Huber PE, Kuner T, von Deimling A, Wick W, Winkler F. Brain tumour cells interconnect to a functional and resistant network. Nature, 2015
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Ç, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SM, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging, 2015
Funding
We are supported by the SFB-824, Deutsche Krebshilfe, TUM-KKF, ZD.B, BMBF, DFG and NIH.